Test de permutación de un score con permutation_test_score — 7:34#
Ultima modificación: 2023-02-27 | YouTube
Evalua la significancia de un score computado con validación cruzada usando permutaciones.
Se permuta la variable de salida para generar datos aleatorios y computar un valor p empírico en contra de la hipótesis nula de que las variables de entrada y la salida son independientes.
Datos#
[1]:
from sklearn.datasets import load_iris
iris = load_iris()
X = iris.data
y = iris.target
Experimento 1#
[2]:
from sklearn.model_selection import StratifiedKFold, permutation_test_score
from sklearn.svm import SVC
clf = SVC(kernel="linear", random_state=7)
cv = StratifiedKFold(2, shuffle=True, random_state=0)
score, permutation_scores, pvalue = permutation_test_score(
# -------------------------------------------------------------------------
# The object to use to fit the data.
estimator=clf,
# -------------------------------------------------------------------------
# The data to fit.
X=X,
# -------------------------------------------------------------------------
# The target variable to try to predict in the case of supervised learning.
y=y,
# -------------------------------------------------------------------------
# Labels to constrain permutation within groups, i.e. y values are permuted
# among samples with the same group identifier. When not specified,
# y values are permuted among all samples.
groups=None,
# -------------------------------------------------------------------------
# Metric
scoring="accuracy",
# -------------------------------------------------------------------------
# Determines the cross-validation splitting strategy.
cv=cv,
# -------------------------------------------------------------------------
# Number of times to permute y.
n_permutations=1000,
# -------------------------------------------------------------------------
# Pass an int for reproducible output for permutation of y values among
# samples.
random_state=0,
# -------------------------------------------------------------------------
# The verbosity level.
verbose=0,
# -------------------------------------------------------------------------
# Parameters to pass to the fit method of the estimator.
fit_params=None,
)
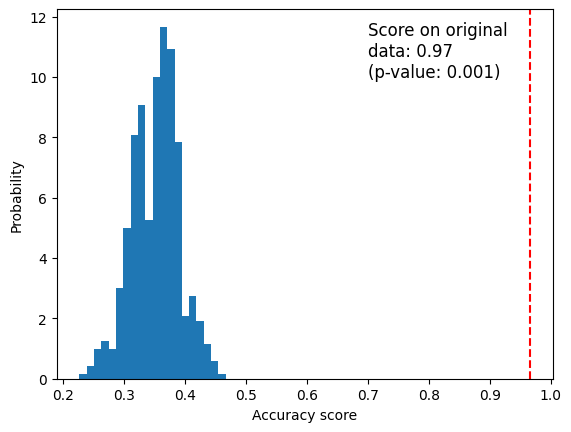
score
[2]:
0.9666666666666667
[3]:
permutation_scores[:10]
[3]:
array([0.33333333, 0.26 , 0.35333333, 0.37333333, 0.33333333,
0.36666667, 0.34 , 0.38 , 0.36666667, 0.34666667])
[4]:
pvalue
[4]:
0.000999000999000999
[5]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.hist(permutation_scores, bins=20, density=True)
ax.axvline(score, ls="--", color="r")
score_label = f"Score on original\ndata: {score:.2f}\n(p-value: {pvalue:.3f})"
ax.text(0.7, 10, score_label, fontsize=12)
ax.set_xlabel("Accuracy score")
ax.set_ylabel("Probability")
plt.show()
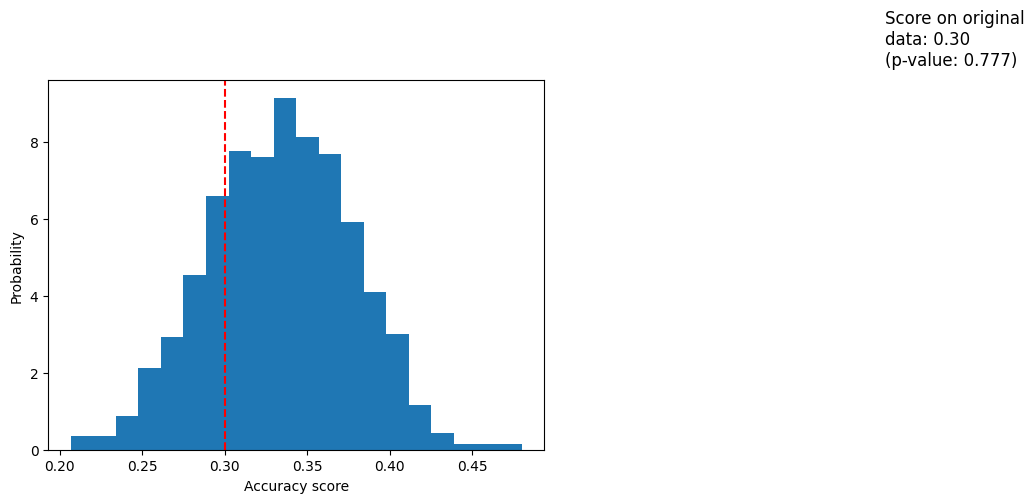
Experimento 2#
[6]:
import numpy as np
n_uncorrelated_features = 20
rng = np.random.RandomState(seed=0)
X_rand = rng.normal(size=(X.shape[0], n_uncorrelated_features))
[7]:
score, permutation_scores, pvalue = permutation_test_score(
clf,
X_rand,
y,
scoring="accuracy",
cv=cv,
n_permutations=1000,
)
score
[7]:
0.30000000000000004
[8]:
permutation_scores[:10]
[8]:
array([0.39333333, 0.28666667, 0.34 , 0.28 , 0.32 ,
0.28 , 0.33333333, 0.31333333, 0.34 , 0.29333333])
[9]:
pvalue
[9]:
0.7772227772227772
[10]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
ax.hist(permutation_scores, bins=20, density=True)
ax.axvline(score, ls="--", color="r")
score_label = f"Score on original\ndata: {score:.2f}\n(p-value: {pvalue:.3f})"
ax.text(0.7, 10, score_label, fontsize=12)
ax.set_xlabel("Accuracy score")
ax.set_ylabel("Probability")
plt.show()